Sedoheptulose-1,7-bisphosphatase (PDB ID:3RPL) from Synechocystis sp. PCC 6803
Created by: Eunice Pyon
Sedoheptulose-1,7-bisphosphatase (PDB ID = 3RPL), also known as SBPase, is a hydrolase found in the cyanobacteria Synechocystis. It is part of the class 1 FBPase family, which is responsible for regulating various metabolic pathways, specifically gluconeogenesis and the Calvin cycle, of different organisms. This family characteristically requires metal ion binding to carry out its catalytic function and is greatly inhibited upon binding with a lithium ion (2).

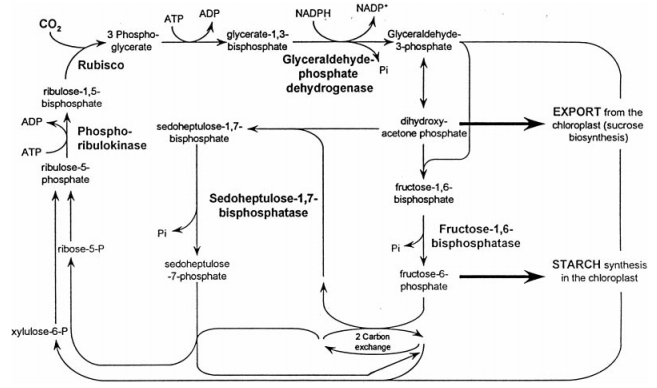
Calvin Cycle (1)
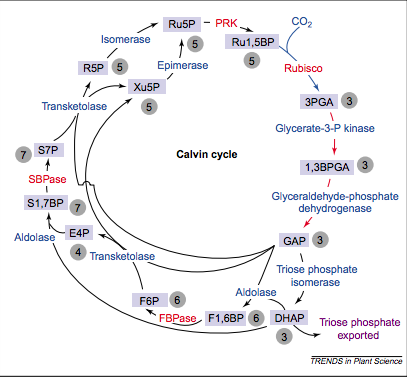
Another Schematic of the Calvin Cycle (13)
SBPase functions as a critical regulator of the carbon flux in the Calvin Cycle, which can be attributed to the correlation between its activity and the amount of carbon fixation. It is a nuclear encoded enzyme that is unique to the Calvin Cycle, with similar functions absent in enzymes of organisms that do not undergo photosynthesis. SBPase is synthesized as a precursor protein with a transit peptide that serves to direct the enzyme to the chloroplast, and is then dissociated (8).
Localized in the thylakoid membrane on the side facing the stroma, sedoheptulose-1,7-bisphosphatase functions as an enzyme catalyst in the dephosphorylation of sedoheptulose-1,7,biphosphate to sedoheptulose-7-phosphate – an event that occurs during the regenerative phase of the Calvin Cycle (1). The photosynthetic carbon dioxide assimilation reaction is irreversible, causing the Calvin Cycle to be restricted to proceeding in the direction of pentose phosphates. This allows for a strict direction in the regeneration of ribulose-1,5-bisphosphate for the initial carboxylation reaction during the Calvin Cycle (6).
SBPase exists as a homodimeric enzyme, consisting of two identical subunits that are approximately 35 kDa each. One subunit contains three polypeptide chains, A, C, and D, and the other subunit contains one polypeptide chain, B. The secondary structure of sedoheptulose-1,7-bisphosphatase consists of 14 α-helices and 18 antiparallel β-strands (12). Two redox-sensitive cysteine residues, Cys52 and Cys57, are located on the regulatory nucleotide-binding domain, forming a disulfide bond that connects the nucleotide domain to the carbon substrate domain (1).
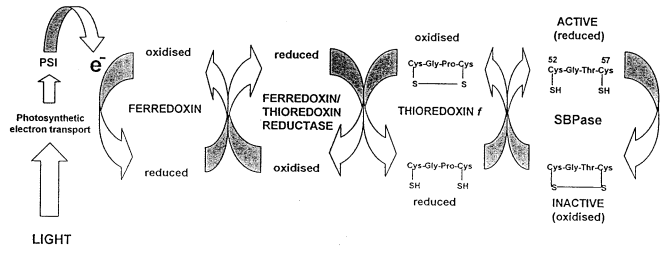
Ferredoxin/thioredoxin-mediated light regulation of SBPase activity (1)
SBPase is inactive in dark conditions, but is activated shortly after illumination. It has been proposed that thioredoxin first forms a stable protein complex with the target enzyme, which produces a reduced SBPase and an oxidized thioredoxin. Thus, the light reactions of photosynthesis generate reducing power that allows ferredoxin and thioredoxin f to reduce the redox-sensitive disulphide bond to two thiol groups, causing a conformational change within the active site of SBPase (3).
SBPase forms complexes with ribulose-5-phosphate kinase (PRKase), glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and ferredoxin reductase, which is an electron acceptor (1). It also necessarily binds to the ligand Mg2+ metal ion to become active, with each subunit binding up to 3 Mg2+ ions (4).
The primary structure of SBPase consists of 393 amino acid residues comprising its four chains, with a total molecular weight of 42414.3 Da and a theoretical isoelectric point of 6.17. Assuming all pairs of Cysteine residues form cystines, the extinction coefficient is 30745 M-1 cm-1. Using the bioinformatics tool BLAST, which uses the FASTA sequence of a desired protein to find proteins with primary sequence homology to a protein query, fructose-1,6-bisphosphatase isolated from Trypanosoma brucei (NCBI ID = Q95PL6) received an E-value of 7e-81 with a 44% sequence identity (11). Previous comparison of E-values from Trypanosoma brucei SBPase and FBPase have indicated that both are targeted to glycosomes. Functional capabilities could be deduced from the sequence homology that exists between the Synechocystis SBPase and the Trypanosoma brucei SBPase (5).
SBPase has several ligands and prosthetic groups in addition to the magnesium ion, which include AMP and beta-fructose-1,6 diphosphate (FBP). SBPase forms a complex with AMP and FBP, in which AMP serves as an allosteric inhibitor and FBP serves as a substrate. AMP binds to the allosteric sites near the interface of C1C4 and C2C3, which are up-down subunit pairs located in the center of the tetramer. Opposite to the interface between C1C2 or C3C4, which are horizontal subunit pairs, is the site of binding of FBP (7).
One of the homologs of SBPase is the enzyme fructose-1,6-bisphosphatase (pub ID = 1FBP), also known as FBPase, isolated from the organism Cupriavidus taiwanensis with an overall 29% sequence identity. The DALI Server, which compares secondary and tertiary structures of proteins, showed FBPase to have a Z-score of 9.0 (14). The hexahedron structure of FBPase is comprised of α-helices and β-sheets linked by turns and loops, which is largely similar to the structure of SBPase. Because of the structural similarity of FBPase to SBPase, functional similarities may also be suggested. Mutagenesis of a wheat cDNA clone has suggested that Cys52 and Cys57 are the two redox-sensitive residues located at the active site. These two residues are located “in a flexible loop near to the junction between the subunits” (1).
Sequence alignment of SBPase in Arabidopsis thaliana has revealed “two GATA motifs with homology to the I box, a conserved DNA sequence element found in the promoter regions of several light-regulated genes” located upstream of the sequence (1). The sequence responsible for regulating gene expression of light-activating portions of SBPase is present in the Chlamydomonas reinhardtii SBPase gene. The SBPase genes in the wheat clone and Arabidopsis show similarities in their intron and exon structures with six introns having the same location (1).
Research is currently being done on sedoheptulose-1,7-bisphosphatase as a potential herbicide target. Lower levels of SBPase and FBPase, in comparison to other enzymes that function in the Calvin Cycle, in transgenic plants have shown a reduced capacity to undergo photosynthesis. This signifies that this reduction in photosynthesis occurs as a result of a reduction in the events of the regenerative phase during the Calvin Cycle (9). Because SBPase is unique to higher level plants, it is a good potential target in herbicide research. There is currently no crystal structure available for SBPase (10).