Overall Reaction of Phospholipase C Delta-1 (PDB ID = 1MAI)
Phospholipase C Delta-1
Created by Jennifer Chan
Phospholipase C Delta-1 (PDB ID = 1MAI) is a
signal transduction protein that does not have alternative conformations or multiple oligomeric states (1). Phospholipase C Delta-1 contains only one subunit,
Chain A, which is composed of 131 residues. Further structural analysis of the protein shows a
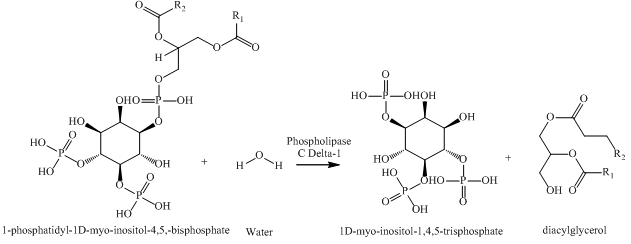
secondary structure displaying 5 alpha helices (shown as red), beta sheets with 7 beta strands (yellow), and 2 random coils and various turns (purple). In addition, Phospholipase C Delta-1 does not contain any protein-drug complex or prosthetic groups. As a whole, the function of Chain A is to aid in the hydrolysis of 1-phosphatidyl-1D-myo-inositol-4,5-bisphosphate (PIP2), a derivative of a phosphatidylinositol, into its secondary messengers (2,3). The two secondary messengers are 1D-myo-inositol-1,4,5-trisphosphate (IP3) and diacylglycerol (DAG) and are involved in signal transduction in cells. More specifically, they result in the release of Ca+2 and the activation of protein kinase C, which in turn, functions to alter cell activity and lead to several physiological changes including tumor production and changes in taste. The overall chemical reaction of this process can be seen below (4).
|
|
|
Overall Reaction of Phospholipase C Delta-1 (PDB ID = 1MAI) |
Although Phospholipase C Delta-1 does not have any directly associated metal ions, it does have an associated ligand, D-myo-inositol-1,4,5-trisphosphate (I3P) (1). I3P allows the protein to attach to its substrate, PIP2, through hydrostatic interactions (3). The protein then cleaves PIP2 into its products, IP3 and DAG, which proceed to function in signal transduction (5). This
binding of a substrate via an associated ligand occurs at the Plextrin Homology (PH) domain (6). This domain, located at the N-terminus of the protein, consists of 119 residues at positions 12-130 and is essential to the role of the protein (7). The
polarities of PH domain residues allow for the protein-lipid interactions (yellow = hydrophobic, pink = polar, blue = charged (+), red = charged (-)) required to bind to lipid vesicles that contain PIP2 (5). In particular, the PH domain of the protein contains several polar and positively charged residues that function in electrostatic interactions with the membrane surface of lipid-containing vesicles to bind them (3). This allows for the hydrolysis of PIP2 into IP3 and DAG and the proper distribution of cellular signals. This collection of residues can be seen in a close-up view of the domain. Additionally, there are
several residues that are functionally important as they come in direct contact with I3P at its binding site. The nine residues are: K30, K32, W36, R40, E54, S55, R56, K57, and T107. These residues are held together to the ligand through hydrogen bonds (dotted spheres). More specifically, the two lysine side chains in K30 and K57 attach themselves directly to the 4- and 5- phosphates of I3P through hydrogen bonding while several other residues link to the 4- and 5- phosphates through direct and water-mediated hydrogen bonds.
Another protein that is structurally similar to Phospholipase C Delta-1 is Engulfment and Cell Motility Protein 1, also known as
ELMO1 (PDB ID = 2VSZ) (8). ELMO1 has two subunits, Chain A and B, and contains 147 residues(1). The secondary structure of ELMO also consists of mainly beta sheets and alpha helices with only a few random coils. Furthermore, not only does ELMO1 (blue) have 78%
similarity in tertiary structure to Phospholipase C Delta-1 (red), but it also contains a PH domain (1,9). Interestingly however, unlike the PH domain of Phospholipase C Delta-1 (which is located at the N-terminus and is able to bind phosphoinositols), the PH domain of ELMO1 is found at the C-terminus and does not have phospholipid-binding properties (9). Thus, although the domains may be similar, ELMO1 does not serve the same biological functions as Phospholipase C Delta-1. Rather, ELMO1 regulates cell motility and phagocytosis of apoptotic cells. Particularly, ELMO1 interacts with another protein, DOCK180 (PDB ID = 3L4C), to maintain proper cell shape and regulate apoptosis in cells (1,10). Despite lacking information regarding this specific interaction, disruptions in ELMO1 are known to lead to improper functioning of DOCK180 and thus a disturbance in the performance of the cell (9).
Compared to many other proteins and enzymes, the structure of Phospholipase C Delta-1 is relatively simple as it only contains one subunit, one ligand, and less than 150 residues. Nonetheless, its structure still allows the protein to fulfill its essential biological functions. The PH domain and its ligand, I3P, play a critical role the binding of phospholinositols and the implementation of cell signal transduction. It is apparent from the physiological form of Phospholipase C Delta-1 that structure and function are complementary and closely linked.