Pyruvate Phosphate Dikinase (PDB ID: 1VBH) from Zea mays
Created by: E. Pierce
Protein 1VBH is pyruvate phosphate dikinase (PPDK), a transferase isolated from Zea Mays (Maize). The enzyme was crystallized with phosphoenolpyruvate (PEP), magnesium, and sulfate bound. PEP is a product of the reaction catalyzed by the enzyme and magnesium interacts in the binding site of PEP (8). 1VBH is a globular enzyme that forms a homodimer. It reversibly catalyzes the transfer of a phosphate group from adenosine triphosphate (ATP) onto pyruvate. It is a dikinase, so it uses ATP to transfer phosphate groups onto two different acceptors (8). This is a key enzyme in photosynthetic pathways, creating PEP. This reaction is key in C4 photosynthesis, which occurs in tropical plants. When the enzyme is found in plants, PEP formation is favorable. In glycolysis it can catalyze the reverse reaction, and create ATP from PEP and pyrophosphate (4). When homologs of the enzyme are isolated in bacteria, the formation of ATP is favored (4).
The enzyme binds ATP in the nucleotide binding region (N terminal region) and pyruvate/ (PEP) in the pyruvate/PEP-binding domain (C terminal region). In the reaction, Histidine residue 458 located in the central domain gains a pyrophosphate group from ATP bound near the N terminus, producing adenosine monophosphate (AMP). A free phosphate ion then accepts one phosphate of the His-Pyrophosphate complex, releasing pyrophosphate. The other phosphate is transferred to a molecule of pyruvate, bound near the C terminus (8). 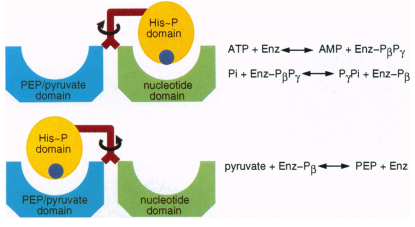 (2). 1VBH was crystallized with and without PEP bound to try to elucidate the swivel motion of the central domain. However the rotation was deduced from comparing protein from maize and C. Symbiosum. In maize the central domain is closest to the PEP binding domain, in C. Symbiosum it is closer to N terminal nucleotide binding site. With the C terminal domain as a reference, the N terminal domain showed little movement. This indicates a rotation of at least 92? of the central domain is required for transfer of the phosphate ion (8).
(2). 1VBH was crystallized with and without PEP bound to try to elucidate the swivel motion of the central domain. However the rotation was deduced from comparing protein from maize and C. Symbiosum. In maize the central domain is closest to the PEP binding domain, in C. Symbiosum it is closer to N terminal nucleotide binding site. With the C terminal domain as a reference, the N terminal domain showed little movement. This indicates a rotation of at least 92? of the central domain is required for transfer of the phosphate ion (8).
 (2). 1VBH was crystallized with and without PEP bound to try to elucidate the swivel motion of the central domain. However the rotation was deduced from comparing protein from maize and C. Symbiosum. In maize the central domain is closest to the PEP binding domain, in C. Symbiosum it is closer to N terminal nucleotide binding site. With the C terminal domain as a reference, the N terminal domain showed little movement. This indicates a rotation of at least 92? of the central domain is required for transfer of the phosphate ion (8).
(2). 1VBH was crystallized with and without PEP bound to try to elucidate the swivel motion of the central domain. However the rotation was deduced from comparing protein from maize and C. Symbiosum. In maize the central domain is closest to the PEP binding domain, in C. Symbiosum it is closer to N terminal nucleotide binding site. With the C terminal domain as a reference, the N terminal domain showed little movement. This indicates a rotation of at least 92? of the central domain is required for transfer of the phosphate ion (8). The protein consists of an 876 amino acid monomer, which dimerizes in solution. The molecular weight is 95192.9 Daltons and its theoretical isoelectric point is 5.27 (3). The protein consists of 46% helical secondary structure and 16% beta sheet structure (6). The protein can be divided into three domains, the N terminal domain, the central domain, and the C terminal domain. The N terminal domain binds ATP, while the C terminal domain binds PEP. The binding sites are 45 angstroms apart. The central domain is responsible for the transfer of a phosphate group by a hypothesized swivel motion (8).
The N terminal domain consists of residues 3 to 343 and contains the nucleotide-binding site. It adopts an ATP grasp fold that consists of 3 subdomains. The first is a beta sheet domain, consisting of residues 3-117, 200-244, and 282-286. This includes a five-stranded antiparallel beta sheet. The second subdomain is a four-helix bundle is inserted into the first subdomain. This helix consists of residues 118-199. The third subdomain is another beta sheet structure, consisting of residues 245-281 and 287-344. It includes another five-stranded antiparallel beta sheet. This ATP grasp fold is well conserved between PPDK’s from different organisms. Magnesium helps bind the ATP molecule (8).
The central domain is the swivel region, which contains the histidine residue that is responsible for the phosphate group transfers(His458). This domain has an alpha/beta sandwich typology, with two beta layers and one layer of alpha helices. The domain is located very close to the C terminal domain, which is distinctly different from other PPDK’s, where the central domain lies closer to the N terminal domain (8). The swivel motion of this domain was determined from comparisons between Clostridium Symbiosum PPDK and Maize PPDK. The central domain is moved by rotation as a rigid body with few conformational changes within the domain.
The C terminal consists of residues 536-876 that contains the pyruvate/PEP binding site. It adopts a TIM barrel motif consisting of eight alpha helices and eight parallel beta strands, with three inserted domains. The inserted region corresponds to residues 566-587, 622-678, and 772-812. The binding site of PEP is located at the end of the beta strands that form the complex inside the barrel. The bind site interacts with PEP, two molecules of sulfate, and a magnesium ion. The magnesium ion coordinates with Glu750 and Asp774. The sulfate ions associate with Arg564, Arg621, and Asn542. The PEP molecule interacts with Arg564, Arg621, Thr772, Asn773, and Asp774. These residues are conserved between other PPDK’s from various organisms. The topology of this region is identical to PPDK’s from other organisms, such as clostridium symbiosum (8).
Linking the central domain to the N terminal domain is a helix-turn-helix motif and a loop consisting of residues 344-393. The two helices have many interactions, and the other loop forms a 3/10 helix. Linking the central domain to the C terminal domain is a loop and a helix consisting of residues 515-535. This linker domain interacts strongly with the first linker domain (8). These linker domains act as the swivel to move the central domain between the two catalytic sites. To show this, the linker domains of CSPPDK and maize PPDK were super imposed. The loops were displaced while the helices stay constant, suggesting conformation changes in the loops are responsible for the rotation of the central domain.
The conformation of the enzyme changes upon binding PEP. The changes are transmitted through the link between the C terminal domain and the central domain. PEP changes the conformation of the side chain of Cys836, located right next to the linker region. This causes Glu838 to change its conformation and interactions; now it interacts with Arg863, which previously interacted with Asp377 and His 381. The center of rotation for the swivel motion is His381. Since Arg863 is no longer interacting with His381, it can move freely. This shows that the binding of PEP to the C terminal domain is the trigger for the swiveling motion of the central domain (8).
1VBH is often compared to the PPDK from Clostridium Symbiosum (1DIK) (7). From the NCBI blast searches and the DALI server, it is shown that the PPKD from C. Symbiosum shares 53% identity with 1VBH, with an E value of 0 and a z score of 49.2 (5,1). In C Symbiosum, this enzyme catalyzes the formation of ATP and pyruvate from PEP and pyrophosphate. The key transfer of the phosphate group still occurs via a key histidine residue. However, in 1DIK, the histidine is located at position 455 (7). The 1DIK monomer is two amino acids smaller than 1VBH, with a total length of 874 amino acids. The monomers exist in solution as homodimers (7). The monomer is still associated into three domains, the N terminal domain, the central domain, and the C terminal domain. The N terminal domain forms the ATP binding site, and carries out the ATP to AMP half of the reaction. The central domain contains His455 and acts as a shuttle, carrying the phosphate group to pyruvate, which is bound in the C terminal domain. This domain catalyzes the formation of PEP. The catalytic sites are approximately 45 angstroms apart, which means that the central domain of 1DIK also must swivel between two conformations. The swivel is approximately 100? around residue 380 (2) Magnesium interacts in the binding sites of the PEP and ATP.
It is interesting to note that the conformational differences between 1DIK and 1VBH lead to the hypothesis of the swiveling central domain mechanism. 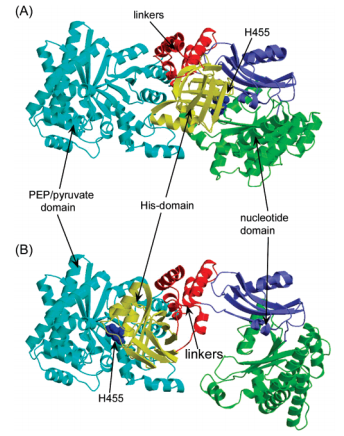 (7). In 1DIK, the central domain is close to the N terminal domain, while in 1VBH it is closer to the C terminal domain. This suggested that the central domain could swivel between the other two domains to transfer a phosphate ion (7, 8).
(7). In 1DIK, the central domain is close to the N terminal domain, while in 1VBH it is closer to the C terminal domain. This suggested that the central domain could swivel between the other two domains to transfer a phosphate ion (7, 8).
 (7). In 1DIK, the central domain is close to the N terminal domain, while in 1VBH it is closer to the C terminal domain. This suggested that the central domain could swivel between the other two domains to transfer a phosphate ion (7, 8).
(7). In 1DIK, the central domain is close to the N terminal domain, while in 1VBH it is closer to the C terminal domain. This suggested that the central domain could swivel between the other two domains to transfer a phosphate ion (7, 8). NCBI protein blasts and the DALI server yielded other close homologs of 1VBH. Another protein that shares sequence identity with PPKD is pyruvate orthophosphate dikinase from P. Paniculatum, with an E value of 0 (1). Other proteins share structural similarities with PPKD. Glycosomal pyruvate phosphate dikinase from trypanosoma brucei had a z score of 50.8 and shared a 54% ID (10, 5). This enzyme favors the reverse reaction, forming ATP in glycolysis. PEP-protein phosphotransferase (Enzyme I) found in E. Coli had a Z score of 32.4 and a 26% ID (5). This enzyme is a transferase found in the bacterial phosphotransferase system (9). Other close structural homologs are more phosphotransferases and dikinases.